All content on this site is intended for healthcare professionals only. By acknowledging this message and accessing the information on this website you are confirming that you are a Healthcare Professional. If you are a patient or carer, please visit the MDS Alliance.
The mds Hub website uses a third-party service provided by Google that dynamically translates web content. Translations are machine generated, so may not be an exact or complete translation, and the mds Hub cannot guarantee the accuracy of translated content. The mds and its employees will not be liable for any direct, indirect, or consequential damages (even if foreseeable) resulting from use of the Google Translate feature. For further support with Google Translate, visit Google Translate Help.
Now you can support HCPs in making informed decisions for their patients
Your contribution helps us continuously deliver expertly curated content to HCPs worldwide. You will also have the opportunity to make a content suggestion for consideration and receive updates on the impact contributions are making to our content.
Find out more
Create an account and access these new features:
Bookmark content to read later
Select your specific areas of interest
View MDS content recommended for you
Editorial theme │ Understanding the new classification and stratification systems in MDS: IPSS-R and IPSS-M
Do you know... The IPSS-M considers bone marrow blasts, platelets, hemoglobin, IPSS-R cytogenetic risk groups, 17 prognostically dominant gene mutations, and number of gene mutations. What is an advantage of the IPSS-M?
The second article in our editorial theme series focuses on understanding the stratification systems in myelodysplastic syndromes (MDS). We focus on the revised International Prognostic Scoring System (IPSS-R) and IPSS-Molecular (IPSS-M). The MDS Hub has previously reported on the development and use of the IPSS-M for MDS risk stratification and prognosis. Real-world data comparing the IPSS-R and the IPSS-M has also been reported on the MDS Hub.
During the European Hematology Association (EHA) 2023 Congress, Hasse presented an overview of MDS risk stratification systems, including the development of the IPSS-R and IPSS-M and their pros and cons.1 Huber presented a poster comparing the IPSS-M and the European LeukemiaNet (ELN) 2022 risk classification.3 In addition, Baer et al.4 recently published a validation study of the IPSS-M in Leukemia. Here, we summarize the key points.
Background to the IPSS1
The IPSS was established in 1997 and incorporates bone marrow blasts, cytopenias, and cytogenetics as relevant prognostic parameters. The IPSS enabled the stratification of prognostic subgroups for overall survival (OS) and acute myeloid leukemia (AML)-free survival; however, it was not an optimal stratification system due to several limitations such as:
- relevance of rare genetic abnormalities is unknown
- arbitrary cytogenetic “intermediate-risk” group
- does not stratify patients with complex abnormalities (≥3 independent clonal anomalies)
- does not consider a combination of chromosomal abnormalities
- based on the small number of patients with an abnormal karyotype
- low prognostic weight of cytogenetics to blasts
- does not consider cytopenias differentially
These limitations led to the development of a cytogenetic prognostic system in 2012, based on an international dataset of 2,902 patients.2 This system included 19 cytogenetic categories (Figure 1) which demonstrated higher prognostic value compared with the previous IPSS (95% vs 81%) and became an integral part of the IPSS-M.
Figure 1. New cytogenetic prognostic system*
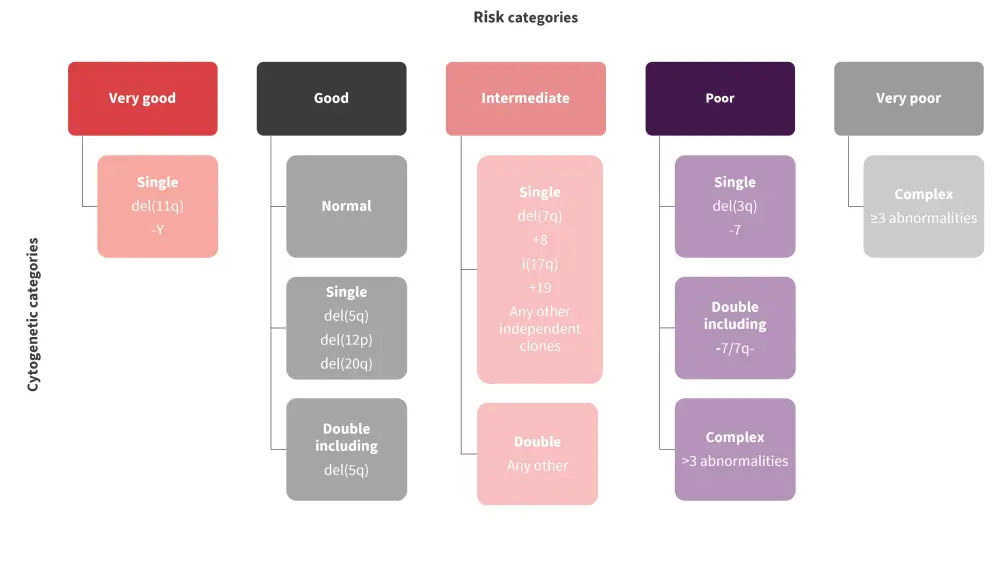
*Adapted from Hasse.1
Objectives of the IPSS-R1
The International Working Group for Prognosis in MDS recognized the limitations of both the IPSS and the cytogenetic prognostic system, and proposed the following revisions:
- validation and implementation of new prognostic parameters such as age and fibrosis;
- integration of the new cytogenetic prognostic system and evaluation of its prognostic impact;
- comprehensive analysis of cytopenias;
- attainment of higher prognostic quality, continuity, applicability, and flexibility; and
- verification of blast thresholds.
The IPSS-R was subsequently developed based on data from 7,012 patients from 18 databases, with a median age of 71 years. The IPSS-R included cytogenetics, bone marrow blasts, hemoglobin, platelets, and absolute neutrophil count as prognostic variables, with very low, low, intermediate, high, and very high prognostic subgroups. Figure 2 lists the pros and cons of the IPSS-R.
Figure 2. Pros and cons of the IPSS-R*
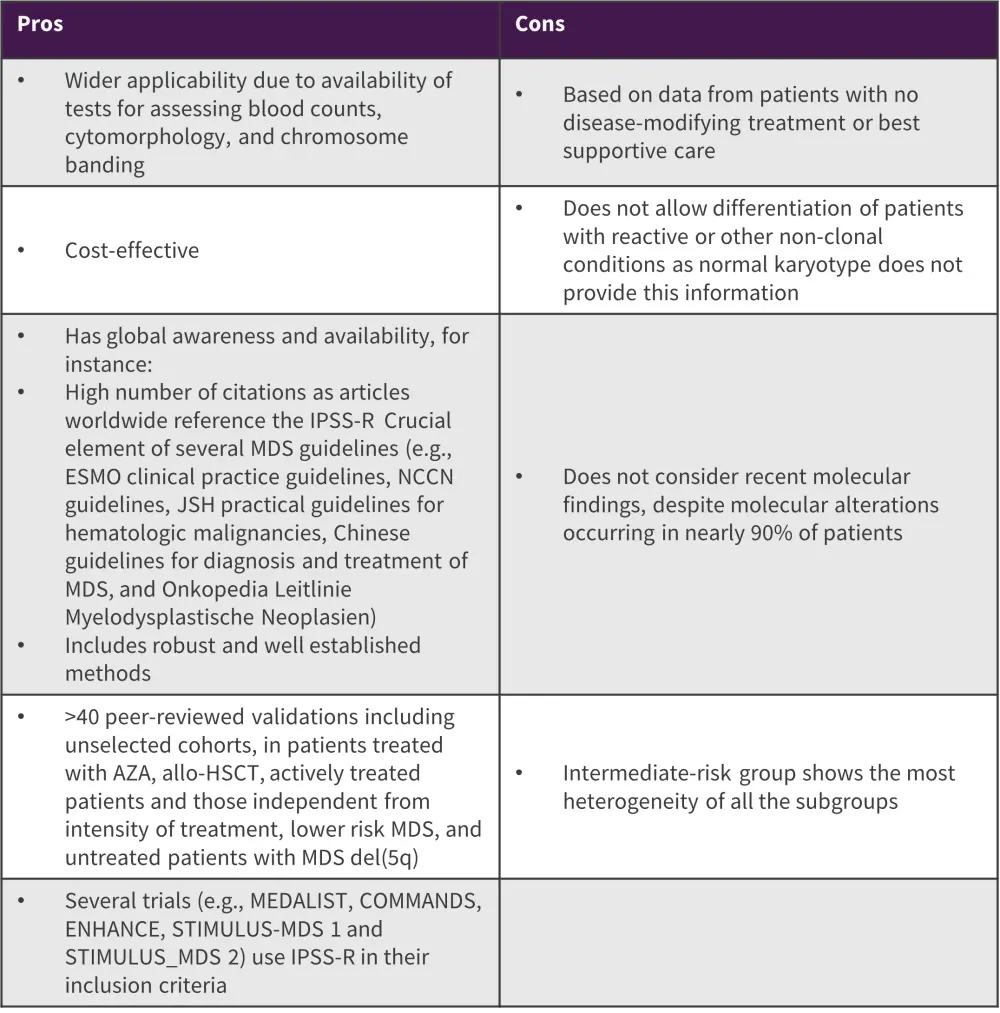
Allo-HSCT, allogeneic hematopoietic stem cell transplantation; Aza, azacitidine; ESMO, European Society for Clinical Oncology; IPSS-R, Revised International Prognostic Scoring System; JSH, Japanese Society of Hematology; MDS, myelodysplastic syndrome; NCCN, National Comprehensive Cancer Network.
*Adapted from Hasse.1
Objectives of the IPSS-M1
With recent findings, adding molecular data to the IPSS-R became important to improve its availability, diagnostic relevance (including proof of clonality in cytopenias and definition of the 5th edition World Health Organization molecularly defined measurable residual disease entities), reproducible molecular consensus profile, pathogenetic meaning, and prognostic impact. Based on these factors, the IPSS-M was proposed to achieve:
- integration of molecular data into the existing risk stratification system
- development of a clinical-molecular prognostic model
- re-evaluation of prognostic relevance, and weight of established and new parameters
- establish an association between parameters and survival outcomes of MDS
- validation of the proposed model using an external cohort
The IPSS-M was based on data from 2,957 patients from 24 centers globally, encompassing 3,073 cytogenetic aberrations and 9,339 mutations across 124 genes. It provides a comprehensive genetic profile of patients with MDS and considers bone marrow blasts, platelets, hemoglobin, IPSS-R cytogenetic risk groups, 17 prognostically dominant gene mutations, and several gene mutations from a panel of 15 genes. The IPSS-M is categorized into six prognostic subgroups of very low, low, moderate low, moderate high, high, high, and very high. The pros and cons of the IPSS-M are shown in Figure 3.
Figure 3. Pros and cons of the IPSS-M*
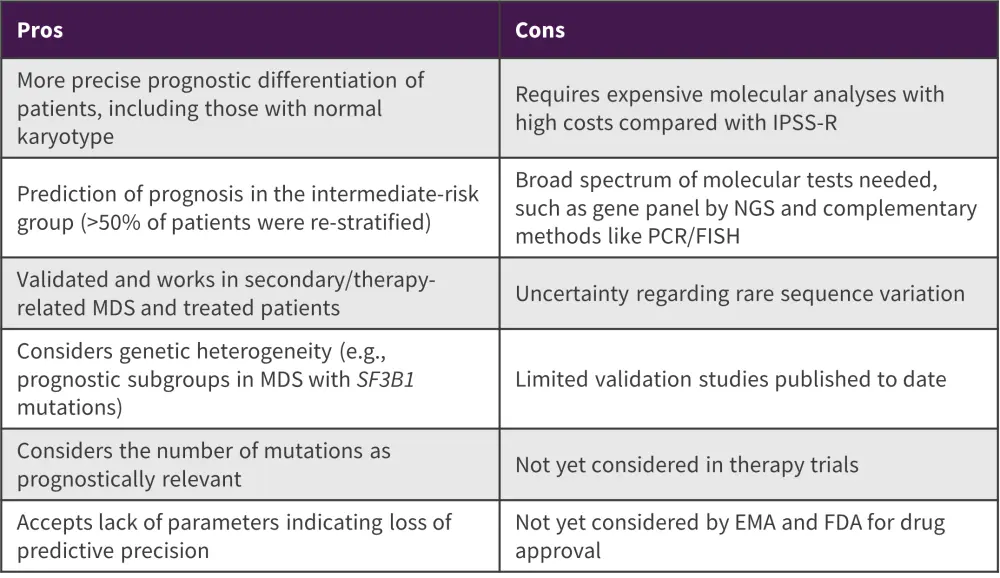
EMA, European Medicine Agency; FDA, Food and Drug Administration; FISH, fluorescent in situ hybridization; IPSS-M, International Prognostic Scoring System-molecular; IPSS-R, Revised IPSS; MDS, myelodysplastic syndrome; NGS, next-generation sequencing; PCR, polymerase chain reaction.
*Adapted from Hasse.1
Comparison of IPSS-M and ELN 2022 risk stratification3
This retrospective cohort study evaluated prognostication in 137 patients with MDS/AML, 626 patients with MDS, and 686 patients with AML using IPSS-M and ELN 2022 risk stratification systems.
IPSS-M risk stratification
- In the MDS/AML cohort, strong prognostic differences were seen in patients with TP53 mutations (14%), myelodysplasia-related gene mutations (72%), myelodysplasia-related cytogenetic abnormalities (4%), or not otherwise specified (10%).
- Most patients with MDS/AML were stratified into high-risk groups, with 45% vs 14% of patients very high-risk, 29% vs 12% high-risk, and 10% vs 7% moderate-high risk compared with the MDS cohort.
- There was clear prognostic separation for OS in the MDS/AML cohort to different IPSS-M risk groups (p < 0.001), with comparable OS in each risk group to the MDS cohort.
ELN 2022 risk stratification
- No patients in the MDS/AML cohort met the criteria for the ELN favorable-risk group.
- Most patients (91%) in the MDS/AML cohort were classified as adverse-risk; these patients had a survival benefit compared with corresponding adverse-risk AML patients (median OS, 1.9 years vs 0.7 years).
- In patients classified as intermediate risk, those in the MDS/AML cohort had an OS benefit over the AML cohort (8.2 years vs 0.8 years).
Validation of IPSS-M4
The real-world cohort study involved data from 626 patients diagnosed with de novo MDS between September 2005 and January 2020, with a median follow-up of 9.5 years. Comparison of the validation cohort with the original publications of the IPSS-M showed that both were comparable in terms of assignment to risk categories (Figure 4) and prognostic separation for OS and leukemia-free survival.5
Individual risk categorization based on both the IPSS-R and IPSS-M in 452 patients showed:
- When IPSS-M moderate-low and moderate-high categories were combined, 25% of patients were upstaged and 19% downstaged by the IPSS-M.
- Risk group differed by one level in 38% of patients, while larger differences (>1 level) level were seen in 6% of patients.
Figure 4. Risk categories in validation vs the original IPSS-M cohort*
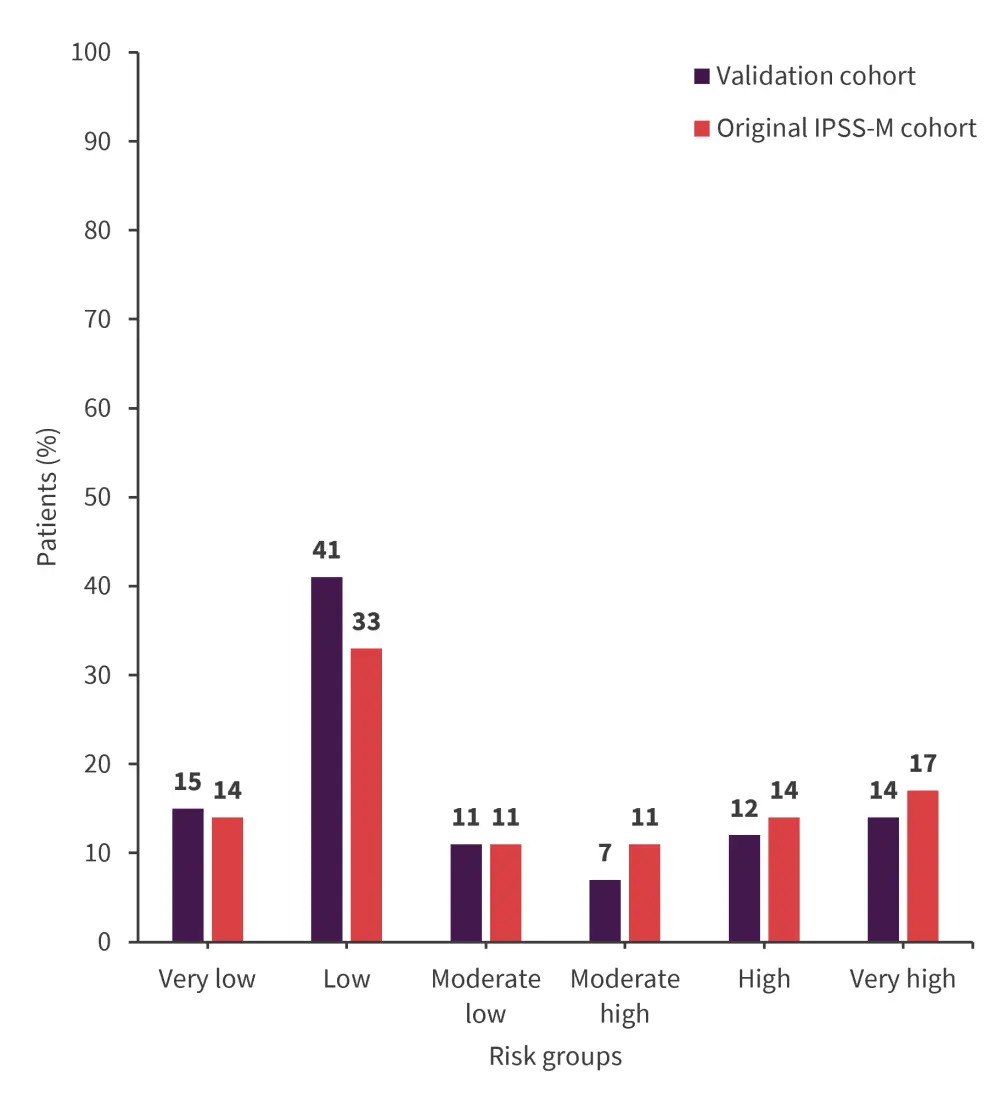
IPSS-M, International Prognostic Scoring System-Molecular.
*Adapted from Baer, et al.4
Harrell’s concordance index to assess the correlation between outcomes based on the IPSS-R and IPSS-M is shown in Figure 5. Age was not considered in either cohort and may have contributed to the lowest OS.
Figure 5. Harrell’s C-index for IPSS-R and IPSS-M*
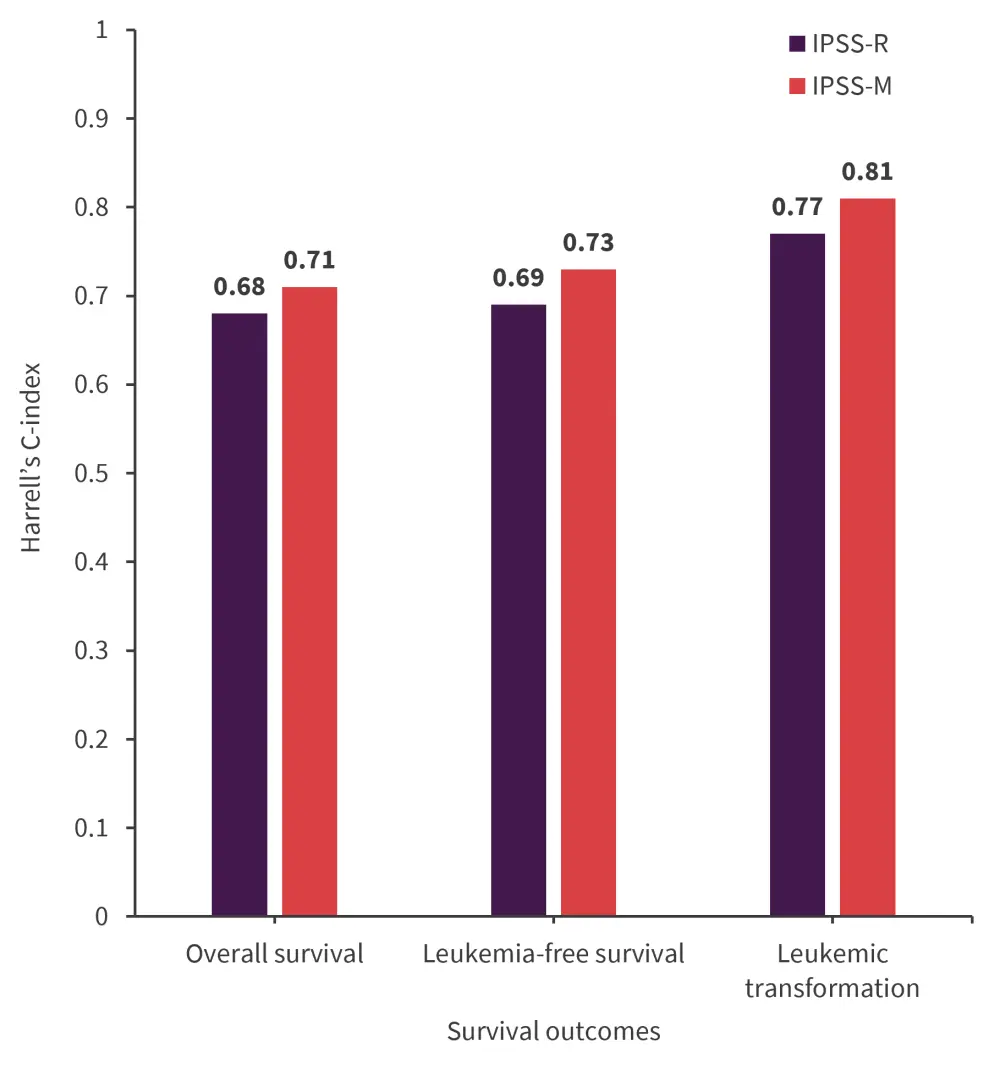
IPSS-M, International Prognostic Scoring System-Molecular; IPSS-R, IPSS-Revised.
*Adapted from Baer, et al.4
The IPSS-M requires 37 parameters in total including TP53 multihit status (TP53multi, combinations of mutations, deletion or copy neutral loss of heterozygosity), KMT2A, FLT3 aberrations, NPM1 mutations, and pattern of co-mutations of SF3B1.
- Within this real-world cohort, 11% of patients had TP53 mutations, of whom 6% were TP53multi.
- Without deletion or copy neutral loss of heterozygosity, one patient would have been missed for TP53multi, suggesting that high variant allele frequency could be a useful surrogate for TP53multi in clinical practice.
- From 199 patients with mutated SF3B1, isolated del(5q) was also present in 9%; whereas, 82% had no specific co-mutations.
- Four patients (one in the very high- and three in the high-risk categories) had mutated NPM1
- One patient remained in the very high-risk group independent of their KMT2APTD status, suggesting determination of KMT2APTD is necessary.
Conclusion
The IPSS-R continues to be the gold standard in MDS prognostication, particularly when the use of the IPSS-M is not possible. Although the IPSS-M provides improved prognostication tools by using validated parameters and up-to-date molecular data, further research is needed to establish its feasibility. However, the ELN 2022 classification does not apply to patients with MDS/AML, and the inclusion of MDS/AML patients in clinical trials assessing adverse risk AML may not be justifiable. Comprehensive molecular analysis is becoming a new standard of MDS prognostication; however, not all laboratories can offer broad molecular genetic panels. As such, laboratories and clinicians must provide a comprehensive set of all types of data in the future, enabling the IPSS-R and the IPSS-M to deliver more meaningful prognostic scores.
References
Please indicate your level of agreement with the following statements:
The content was clear and easy to understand
The content addressed the learning objectives
The content was relevant to my practice
I will change my clinical practice as a result of this content



