All content on this site is intended for healthcare professionals only. By acknowledging this message and accessing the information on this website you are confirming that you are a Healthcare Professional. If you are a patient or carer, please visit the MDS Alliance.
The mds Hub website uses a third-party service provided by Google that dynamically translates web content. Translations are machine generated, so may not be an exact or complete translation, and the mds Hub cannot guarantee the accuracy of translated content. The mds and its employees will not be liable for any direct, indirect, or consequential damages (even if foreseeable) resulting from use of the Google Translate feature. For further support with Google Translate, visit Google Translate Help.
Now you can support HCPs in making informed decisions for their patients
Your contribution helps us continuously deliver expertly curated content to HCPs worldwide. You will also have the opportunity to make a content suggestion for consideration and receive updates on the impact contributions are making to our content.
Find out more
Create an account and access these new features:
Bookmark content to read later
Select your specific areas of interest
View MDS content recommended for you
Targeted sequencing of seven genes may improve classification of MDS
The National Myelodysplastic Syndromes (MDS) Natural History Study (NCT02775383) is a collaborative study involving the National Heart, Lung, and Blood Institute (NHLBI), National Cancer Institute (NCI), and NCI Community Oncology Research Program (NCORP). It is currently investigating whether targeted exon sequencing may be useful to increase compliance between local and central pathologic review of diagnosis, by analyzing the clinical, genetic, and epigenetic characteristics related to the onset and evolution of the disease in patients with newly diagnosed or suspected MDS.
The results were presented by Johannes Goll during the 62nd American Society of Hematology Annual Meeting and Exposition,1 and here, we are pleased to summarize the key points of his talk.
Background
- The National MDS Natural History Study is a prospective, observational cohort study:
- Conducted at > 120 centers
- With an enrolment target of up to 2,000 patients with MDS or MDS/myeloproliferative neoplasms (MPN) overlap, and up to 500 patients with idiopathic cytopenias of unknown significance (ICUS)
- Peripheral blood and bone marrow samples collected from untreated, cytopenic patients and previously examined by local pathologists were reviewed by central pathologists; an independent review was used if there was a conflict
- Diagnosis classifications were as follows:
- MDS*
- MDS/MPN overlap
- Acute myeloid leukemia (AML) blasts < 30%
- Other: clonal cytopenia, morphologic dysplasia, AML blasts ≥ 30%, other myeloid malignancies
- The agreement between local and centralized pathologic review was examined against these diagnostic classifications
- The largest absolute discrepancies were observed between the diagnosis of MDS versus Other
- Therefore, the goal of this study was to assess whether targeted gene sequencing could be used to increase agreement between pathologic diagnoses in these two categories.
*The updated 2016 World Health Organization (WHO) classifications were followed to diagnose MDS in the central review.
Methods
A total of 648 patients (MDS, n = 212; Other, n = 436) were included in the analysis. There were 90 patients with clonal cytopenia of undetermined significance (CCUS) and 89 patients with other cancers in the Other category.
Targeted exon sequencing was performed in 96 myeloid genes with a mean coverage of 1,317X. The threshold for minimum variant allele frequency (VAF) was 2%. To create the model and perform the first test, likely disease-causing variants were manually reviewed in 596 subjects (Figure 1).
Figure 1. Methods1
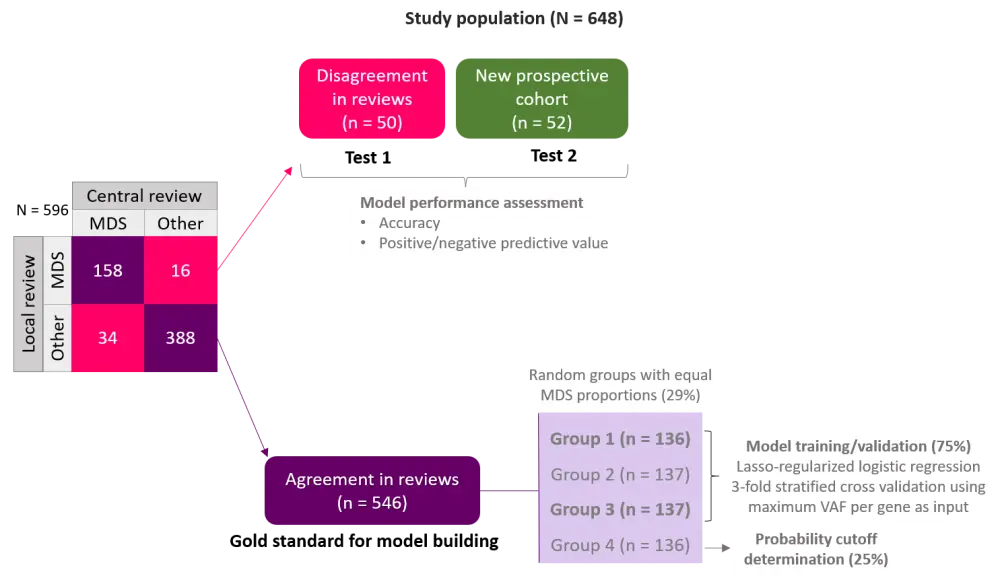
MDS, myelodysplastic syndromes, VAF, variant allele frequency.
Dataset from gold standard group (n = 546) where there was agreement in local and central reviews, was used to build the model and classify genes based on maximum VAF. This model was then tested in the study population where there was disagreement in reviews (Test 1), and prospectively in a group with diagnosis subsequently confirmed by histological assessment (Test 2).
Results
The model building process proposed seven genes as the most informative genes in predicting MDS versus Other, namely TP53, SF3B1, U2AF1, ASXL1, TET2, STAG2, and SRSF2, in order of decreasing impact. The possibility of predicting MDS was increased with increasing maximum VAF for all these genes.
In the determination of probability cutoff point, a cutoff of ≥ 0.17 was identified for predicting MDS versus Other with a sensitivity of 0.90 for true positives and specificity of 0.81 for false positives. The discrimination on the receiver operating characteristic (ROC) curve was satisfactory for the seven genes with an area under the curve (AUC) of 0.89.
The model could decrease the disagreement between local and central reviews by 74% (Table 1) and produced accurate results for the prospective cohort by 83% (Table 2). Gene modelling led to an increase in MDS diagnosis agreed both by local and central reviews compared with traditional classification (Figure 2).
Table 1. Results from Test 11
|
MDS, myelodysplastic syndromes; NPV, negative predictive value; PPV, positive predictive value. |
|||||||||
|
Traditional classification |
|
Gene modelling |
Accuracy: 0.74 |
||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
Central review |
|
Central review |
PPV: 0.89 |
|||||
|
MDS |
Other |
MDS |
Other |
NPV: 0.57 |
|||||
|
Local review |
MDS |
158 |
16 |
Local review |
MDS |
182 |
3 |
Sensitivity: 0.71 |
|
|
Other |
34 |
388 |
Other |
10 |
401 |
Specificity: 0.81 |
|||
Figure 2. MDS diagnosis before and after gene modelling1
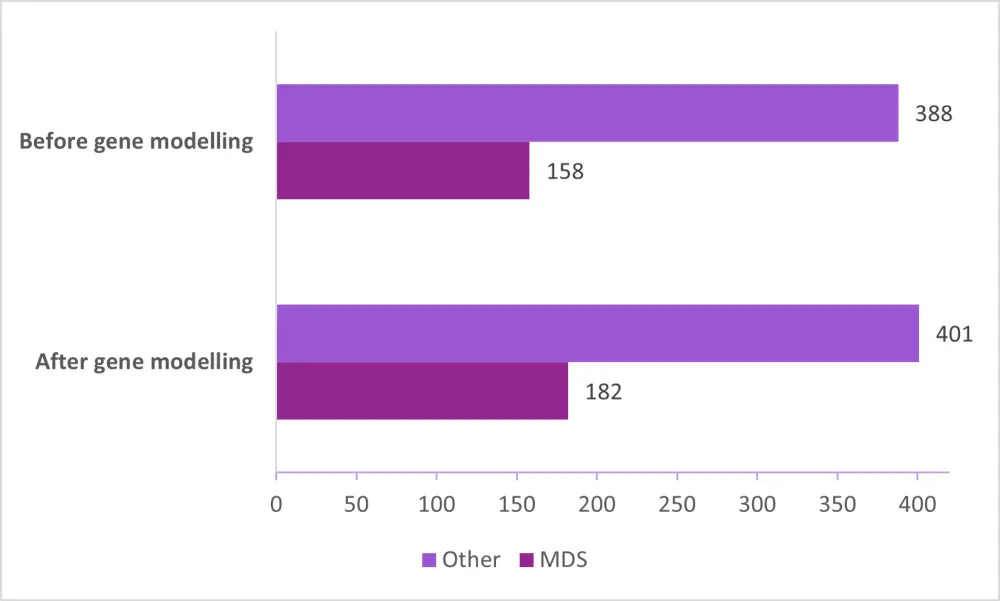
MDS, myelodysplastic syndromes.
Table 2. Results from Test 2 (prospective cohort; n = 52)1
|
MDS, myelodysplastic syndromes; NPV, negative predictive value; PPV, positive predictive value. |
||||
|
|
Final histopathology |
Accuracy: 0.83 |
||
|---|---|---|---|---|
|
PPV: 0.83 |
||||
|
MDS |
Other |
NPV: 0.82 |
||
|
7-gene classifier |
MDS |
15 |
3 |
Sensitivity: 0.83 |
|
Other |
6 |
28 |
Specificity: 0.82 |
|
Conclusion
The use of targeted exon sequencing allowed reclassification of subjects diagnosed with MDS or Other in 74% occurrences of disagreement between local and central pathology reviews. The evaluation of an independent, prospective cohort produced results with an accuracy of 83%. Overall, study results suggest that using targeted sequencing of seven genes may improve the classification of MDS in cytopenic patients.
References
Please indicate your level of agreement with the following statements:
The content was clear and easy to understand
The content addressed the learning objectives
The content was relevant to my practice
I will change my clinical practice as a result of this content

