All content on this site is intended for healthcare professionals only. By acknowledging this message and accessing the information on this website you are confirming that you are a Healthcare Professional. If you are a patient or carer, please visit the MDS Alliance.
The mds Hub website uses a third-party service provided by Google that dynamically translates web content. Translations are machine generated, so may not be an exact or complete translation, and the mds Hub cannot guarantee the accuracy of translated content. The mds and its employees will not be liable for any direct, indirect, or consequential damages (even if foreseeable) resulting from use of the Google Translate feature. For further support with Google Translate, visit Google Translate Help.
Now you can support HCPs in making informed decisions for their patients
Your contribution helps us continuously deliver expertly curated content to HCPs worldwide. You will also have the opportunity to make a content suggestion for consideration and receive updates on the impact contributions are making to our content.
Find out more
Create an account and access these new features:
Bookmark content to read later
Select your specific areas of interest
View MDS content recommended for you
TP53 allelic state has implications in MDS in terms of genomic stability, clinical presentation, and outcomes
The oncogene tumor protein 53 (TP53) is established as the most commonly mutated gene across cancer types. Within the context of myelodysplastic syndromes (MDS), TP53 mutations are strongly associated with an increased risk of acute myeloid leukemia (AML), more rapid transformation, and aggressive and refractory disease with poor outcomes. Genetic mutations within TP53 are allelic, with both monoallelic and biallelic mutations identified in patients with MDS. What is not known is how different allelic variants translate to clinical outcomes and treatment responses for patients with MDS.1
Here, we report research by Elsa Bernard and colleagues, recently published in Nature Medicine in October 2020, exploring how different allelic states of TP53 affect the clinical presentations and outcomes of patients with TP53-mutated MDS.1
Study design
- An international, multicenter, biological observational cohort study (centres in USA, Europe, Japan, and Brazil).
- Collaboration with the International Working Group for Prognosis in MDS.
Case population
- A total of 3,324 patients were included in the molecular analysis.
- All patients had MDS, with no AML transformation.
Clinical data
The following clinical data were collected:
- Demographic (sex and age at diagnosis)
- MDS data (type, World Health Organization 2016 classification, revised international prognostic scoring system [IPSS-R] risk score, and cytogenetic risk)
- Biomarker profiles (blood count, bone marrow blasts, and ferritin)
- Outcome data (follow-up, overall survival [OS], and AML transformation)
- Treatment (hematopoietic stem cell transplant, lenalidomide, and hypomethylating agents).
Clinical endpoints included length of follow-up, median OS, 3-year OS, 3-year transformation to AML, and time to treatment failure.
Genetic/molecular analysis
Mutational analysis was performed using the following:
- Conventional G-banding analyses
- Tumor-only next generation sequencing
- Genome-wide copy number probes
- Allele-specific copy number analysis.
Results
A total of 378 patients were identified with TP53 mutations, among which:
- 274 patients (72.5%) had single mutations
- 100 patients (26.5%) had two mutations
- 4 patients (1%) had three mutations.
Table 1 presents the 3-year OS and AML risk for patients with TP53 mutations and common MDS chromosomal risk factors.
Table 1. Outcomes for patients with single TP53 mutations
|
Single TP53 mutation, no -17/17p mutation (by cytogenetics), no focal del17p, and no 17/17p cnLOH |
All other patients |
|
|
TP53, tumor protein 53; OS, overall survival; AML, acute myeloid leukemia; cnLOH, copy neutral loss of heterozygosity. |
||
|
Number of patients |
125 |
253 |
|
Median OS, years |
2.5 |
0.7 |
|
3-year OS, % |
49 |
8 |
|
3-year AML risk, % |
16 |
40 |
Two thirds of patients with TP53 mutations had multiple hits, with more than one gene mutation, gene mutation and deletion, or mutation and copy-neutral loss of heterozygosity.
In total, 14 (13%) patients with more than one TP53 mutation had concomitant allelic imbalance at the TP53 locus compared with 149 (54%) patients with one TP53 mutation (OR, 7.6; 95% CI, 4.10–15.20). A higher proportion of poor/very poor IPSS-R patients were seen in the multi-hit group of patients (74%) than in the monoallelic group (9%) (OR, 28.0; p = 2x10‑35 two-sided Fisher’s exact test).
The 5-year survival rates were influenced by co-occurring driver mutations for patients with monoallelic TP53 mutations (Figure 1).
Figure 1. Five-year survival (%) of patients with monoallelic TP53 mutations according to the number of co-occurring genetic mutations
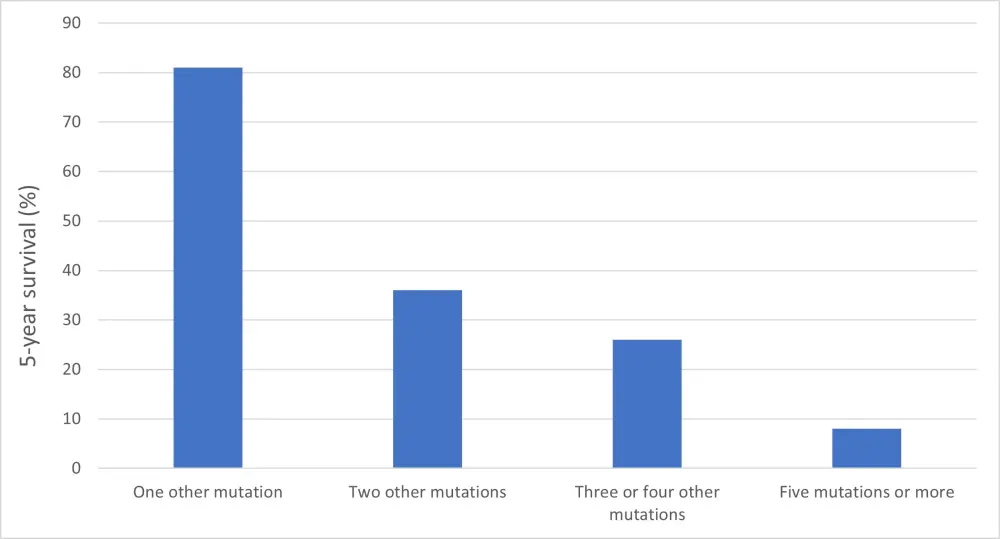
Therapy-related MDS (t-MDS) carried a higher proportion of TP53-mutated patients (18%) relative to patients with de novo MDS (6%) (OR, 3.3; 95% CI, 2.4–4.6), with an increased risk of multiple hits (84% t-MDS vs 65% de novo MDS; OR, 2.8; 95% CI, 1.4–6.6). Monoallelic mutated t-MDS had a lower risk of death than patients with multi-hit t-MDS (HR, 0.39; 95% CI, 0.15–1.00; P=0.05).
Patients with monoallelic TP53 mutations treated with hypomethylating agents and lenalidomide showed a trend to longer survival, as did patients treated with hematopoietic stem cell transplant.
Conclusion
Monoallelic TP53-mutated patients experience significantly different outcomes relative to patients carrying multi-hit TP53 genetic aberrations. Patients with a multi-hit profile demonstrated higher degrees of genomic instability, high-risk disease, treatment refractory disease, and reduced OS. Furthermore, multi-hit TP53 mutations can identify very high-risk patients alongside established poor prognostic markers, such as IPSS-R. There was no difference in OS, AML progression, or treatment response in patients with monoallelic TP53 mutation compared with patients with wild-type TP53.
The authors conclude that given the prevalence of TP53 mutations, and the significance of multi-hit TP53 mutations for adverse outcomes in MDS, TP53 allelic analysis should become a routine part of MDS assessment, classification, staging, and future clinical trials and studies.
References
Please indicate your level of agreement with the following statements:
The content was clear and easy to understand
The content addressed the learning objectives
The content was relevant to my practice
I will change my clinical practice as a result of this content

