All content on this site is intended for healthcare professionals only. By acknowledging this message and accessing the information on this website you are confirming that you are a Healthcare Professional. If you are a patient or carer, please visit the MDS Alliance.
The mds Hub website uses a third-party service provided by Google that dynamically translates web content. Translations are machine generated, so may not be an exact or complete translation, and the mds Hub cannot guarantee the accuracy of translated content. The mds and its employees will not be liable for any direct, indirect, or consequential damages (even if foreseeable) resulting from use of the Google Translate feature. For further support with Google Translate, visit Google Translate Help.
Now you can support HCPs in making informed decisions for their patients
Your contribution helps us continuously deliver expertly curated content to HCPs worldwide. You will also have the opportunity to make a content suggestion for consideration and receive updates on the impact contributions are making to our content.
Find out more
Create an account and access these new features:
Bookmark content to read later
Select your specific areas of interest
View MDS content recommended for you
The impact of CD33 and ABCB1 SNPs on clinical outcomes in AML: A retrospective analysis of a phase II decitabine and gemtuzumab ozogamicin study in AML and high-risk MDS
Single nucleotide polymorphisms (SNPs) of CD33 are predictive of relapse in pediatric patients with acute myeloid leukemia (AML) treated with gemtuzumab ozogamicin (GO) frontline therapy. Likewise, SNPs of ABCB1 do not only impact the accumulation of calicheamicin mediated by GO in preclinical studies, but also are thought to impact the relapse-risk in pediatric patients with AML. The evidence on this impact is not clear in adult patients.
In a phase II study (NCT00882102) investigating the efficacy and safety of the combination of GO and decitabine, an improvement of the response rate, but not overall survival (OS), was observed in treatment-naïve patients with AML (aged ≥ 60 years) who were unsuitable for chemotherapy, AML evolving from treated MDS, and previously treated MDS or myelofibrosis (MF), in comparison to historical controls.1 Recently, a retrospective analysis has been published in American Journal of Hematology by Nicholas Short and colleagues. The authors evaluated the impact of SNPs of CD33 and ABCB1 in patients with AML, high-risk MDS, chronic myelomonocytic leukemia, primary MF, or patients who received the combination of decitabine and GO in a clinical trial (NCT00882102) or settings outside of a clinical trial.2
Study design
Of 113 patients who received decitabine plus GO in a clinical trial setting or outside of a clinical trial,1 a retrospective analysis of stored samples was performed on 104 patients (101 on-protocol; 3 off-protocol):2
- Genotyping of different SNPs was performed and then compared with clinical outcomes: ABCB1 rs1128503, ABCB1 rs1045642, CD33 rs2455069, CD33 rs35112940, CD33 rs12459419, CD33 rs1803254, CD33 rs61736475, and CD33 rs201074739
- An aggregate “6 CD33 SNP score” was determined where a higher score indicated an increased anti-leukemia response to GO
- Mean fluorescence intensity (MFI) was used to evaluate leukemic cell surface expression of CD33
Patient characteristics2
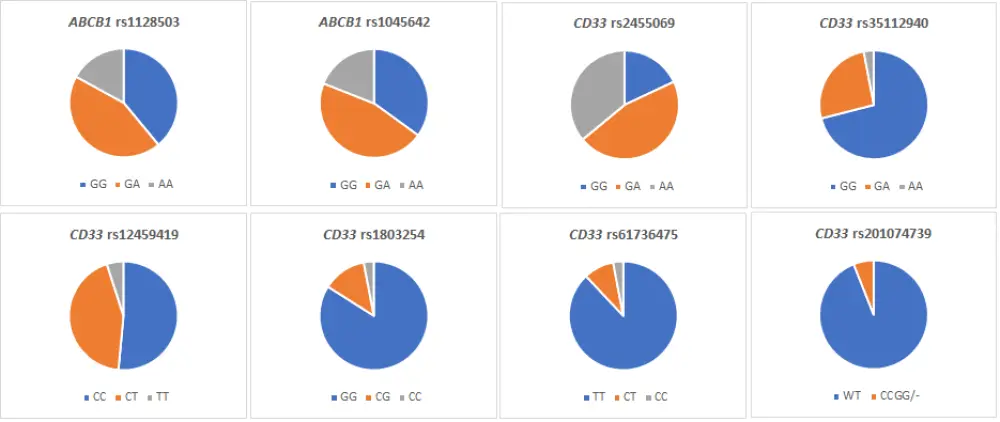
The baseline characteristics of the evaluable patients are shown in Table 1. In Figure 1 we report the frequency of CD33 and ABCB1 polymorphisms.
Table 1. Patient characteristics
|
AML, acute myeloid leukemia; CMML, chronic myelomonocytic leukemia; MDS, myelodysplastic syndromes; MFI, mean fluorescence intensity; PMF, primary myelofibrosis; SNP, single nucleotide polymorphism *Includes patients with MDS (n = 15), CMML (n = 4) or PMF (n = 3) |
|||
|
Characteristic |
Total (n = 104) |
Frontline cohort (n = 49) |
Relapsed/Refractory cohort (n = 55) |
|---|---|---|---|
|
Age, years (range) ≥ 60 years old, n (%) |
68 (24─88) 80 (77) |
70 (42─87) 44 (90) |
65 (24─88) 36 (65) |
|
Diagnosis, n (%) AML MDS/CMML/PMF* |
82 (79) 22 (21) |
36 (73) 13 (27) |
46 (84) 9 (16) |
|
Secondary AML, n (%) |
23/82 (28) |
6/36 (17) |
17/48 (35) |
|
Therapy-related |
31 (30) |
17 (35) |
14 (25) |
|
Median CD33 expression, % (range) |
82.8 (0─100) |
85 (0─100) |
81.9 (8.9─99.6) |
|
Median CD33 MFI ratio, (range) |
29.8 (1─180.4) |
27.1 (1─158) |
30.6 (3.6─180.4) |
|
6 SNP score < 0 ≥ 0 |
51 (49) 53 (51) |
|
|
Figure 1. Frequency of CD33 and ABCB1 polymorphisms
Results2
- Median MFI was not found to be significantly associated with any individual SNP or the 6 CD33 SNP score
- The overall response rate (ORR; composite of complete remission [CR] plus CR with inadequate hematologic recovery plus morphologic leukemia-free state) for the entire cohort was 32% (n = 33)
- The CR rate was 14% (n= 15)
- Clinical outcomes for the frontline and relapsed/refractory cohorts are reported in Table 2
Table 2. Clinical outcomes in the frontline and relapsed/refractory cohorts
|
CR, complete response; ORR, overall response rate; OS, overall survival; RFS, relapse-free survival *Median follow-up period of 107 months |
||
|
Clinical outcomes |
Frontline cohort |
Relapsed/refractory cohort |
|---|---|---|
|
ORR, % |
47 |
18 |
|
CR rate, % |
22 |
7 |
|
RFS*, months |
4.7 |
4.4 |
|
OS*, months |
10.0 |
6.7 |
|
1-year OS, % |
43 |
29 |
|
2-year OS, % |
12 |
13 |
- CD33 MFI was significantly associated with CR both in univariate (odds ratio [OR] 1.02, 95% confidence interval [CI], 1.00─1.03; p = 0.01) and multivariate analysis (p = 0.01), after adjusting for age, diagnosis (AML or others), disease status, and cytogenetics
- No individual CD33 or ABCB1 SNP was predictive for ORR or CR
- A trend towards higher CR rate was observed in patients with 6 CD33 SNP score ≥ 0 vs 6 CD33 SNP score < 0 (21% vs 8%; p = 0.07), however this was not the case for ORR
- There was no significant association between CD33 MFI or 6 CD33 SNP score and cumulative incidence of relapse (CIR), relapse-free survival (RFS), or OS
- Only the CD33 rs1803254 SNP was associated with CIR (p = 0.02) and RFS (p = 0.03), but not with OS (p = 0.11)
- Results from off-protocol patients (n = 3) revealed that the median RFS was longer (5.7 vs 2.5 months; hazard ratio, 4.54, 95% CI ,1.19─17.25; p = 0.03) and CIR was lower (73% vs 100%; p = 0.02) in patients with GG genotypes compared to those with GC/CC genotypes
- For CC/CG and GG genotypes, there were associations between the CD33 rs1803254 SNP and CIR (1.8 vs 10.1 months; p < 0.01), RFS (2.2 vs 6.8 months; p < 0.01), and OS (6.6 vs 11.4 months; p = 0.04) in patients with frontline disease, indicating CC/CG genotype was associated with worse CIR and RFS
- CC or CG genotypes of CD33 rs1803254 were present in 16% of patients, and higher relapse rates were seen in these patients
- However, the impact of CD33 rs1803254 on RFS or OS in patients treated with decitabine plus GO was not significant
Limitations2
The study population had very poor-risk disease and was comparably heterogeneous, which may affect the detection of differences based on genotypes. The ORR with the combination of decitabine plus GO was relatively low, therefore, the number of patients evaluated in CIR and RFS analyses was small.
Conclusion
Only the CD33 rs1803254 GC/CC genotype was associated with significantly worse CIR and RFS in patients diagnosed with AML or other advanced myeloid malignancies, while other genotypes did not show any significant associations with clinical outcomes. In contrast to previous findings reported in children, where the CD33 rs12459419 SNP had been shown to influence the long-term treatment outcomes with chemotherapy plus GO, this study did not demonstrate such an association with the combination of decitabine and GO.2 On the contrary, this finding supports two other analyses which also found no association between the CD33 rs12459419 SNP and the outcomes of patients treated with regimens containing GO or the anti-CD33 antibody drug conjugate SGN-CD33A.3,4 The authors emphasized a higher frequency of CD33 rs1803254 CG/CC genotype in black patients who had worse CIR and RFS outcomes compared to non-black patients (38% vs 13%), however, the small number of patients did not allow a full assessment of the relationship between genotype, race, and poorer outcomes.2
References
Please indicate your level of agreement with the following statements:
The content was clear and easy to understand
The content addressed the learning objectives
The content was relevant to my practice
I will change my clinical practice as a result of this content

