All content on this site is intended for healthcare professionals only. By acknowledging this message and accessing the information on this website you are confirming that you are a Healthcare Professional. If you are a patient or carer, please visit the MDS Alliance.
The mds Hub website uses a third-party service provided by Google that dynamically translates web content. Translations are machine generated, so may not be an exact or complete translation, and the mds Hub cannot guarantee the accuracy of translated content. The mds and its employees will not be liable for any direct, indirect, or consequential damages (even if foreseeable) resulting from use of the Google Translate feature. For further support with Google Translate, visit Google Translate Help.
Now you can support HCPs in making informed decisions for their patients
Your contribution helps us continuously deliver expertly curated content to HCPs worldwide. You will also have the opportunity to make a content suggestion for consideration and receive updates on the impact contributions are making to our content.
Find out more
Create an account and access these new features:
Bookmark content to read later
Select your specific areas of interest
View MDS content recommended for you
Prevalence and impact of PPM1D mutations in patients with MDS and del(5q)
Mutations in the PPM1D gene drive clonal hematopoiesis and occur in various malignancies. Gain-of-function mutations in PPM1D are known to impair TP53 function, and are therefore considered to act in a similar manner to the TP53 loss-of-function mutations that are reported in 5−10% of patients with myelodysplastic syndromes (MDS) and enriched in patients with del(5q).
Victoria Panagiota and colleagues have investigated the prevalence and impact of PPM1D mutations on the outcomes of patients with MDS and del(5q).1 Their study results, published in the American Journal of Hematology, are summarized below.
Study design
A retrospective analysis of 234 adult patients with MDS or secondary acute myeloid leukemia (sAML) and del(5q), subdivided into three groups:
- WHO 2016 defined del(5q) MDS, comprising patients with del(5q) alone or one additional abnormality, excluding monosomy 7 or del(7q), <5% bone marrow blasts and <1% peripheral blood blasts (n = 175)
- Other MDS with del(5q), comprising all other cases of MDS with del(5q) and complex karyotype, chromosome 7 abnormalities or >5% bone marrow blasts or >1% peripheral blood blasts (n = 38)
- sAML with del(5q) (n = 21)
Results
Median age was 72.2 years, and 72.2% of patients were female. Del(5q) was the sole chromosomal abnormality in 86.3% of patients; 4.3% had at least one additional cytogenetic abnormality, and 9.4% had a complex karyotype.
In the overall cohort, 37.1% of patients were treated with lenalidomide, 29% of patients progressed to AML and 8.1% underwent allogeneic hematopoietic stem cell transplantation.
Mutation status
PPM1D and TP53 mutation status at time of diagnosis for the total cohort and according to the three subgroups of patients with overall survival (OS) information, is shown in Table 1.
- Of the 13 patients with mutated PPM1D, one had a trisomy 8, two had a complex karyotype, and three had a TP53 co-mutation (including the two patients with a complex karyotype and one with WHO 2016 defined del(5q) MDS)
- 12 of the 35 patients with mutated TP53 had a complex karyotype
Table 1. Prevalence of PPM1D and TP53 mutations for MDS/sAML patients with del(5q)*
|
Del, deletion; MDS, myelodysplastic syndromes; mut, mutation; OS, overall survival; sAML, secondary acute myeloid leukemia; WHO, World Health Organization; wt, wildtype. |
||||
|
Mutation status, n (%) |
Total cohort |
WHO 2016 defined del(5q) MDS† |
Other MDS with del(5q)† |
sAML with del(5q)† |
|---|---|---|---|---|
|
PPM1Dmut |
13 (5.6) |
11 (6.7) |
2 (6.5) |
0 (0) |
|
TP53mut |
35 (15) |
17 (10.4) |
8 (25.8) |
7 (33.3) |
|
PPM1Dwt/TP53wt |
189 (80.8) |
137 (83.5) |
23 (74.2) |
14 (66.7) |
|
PPM1Dmut/TP53mut |
3 (1.3) |
1 (0.6) |
2 (6.5) |
0 (0) |
|
PPM1Dwt/TP53mut |
32 (13.7) |
16 (9.8) |
6 (19.4) |
7 (33.3) |
|
PPM1Dmut/TP53wt |
10 (4.3) |
10 (6.1) |
0 (0) |
0 (0) |
Prognostic impact of PPM1D mutations
After a median follow-up of 2.6 years, two patients (18.2%) with WHO 2016 defined del(5q) MDS and mutated PPM1D transformed to AML. AML transformation rates were 6.3% and 20.4% for patients with wildtype PPM1D/mutated TP53 and patients with wildtype PPM1D/wildtype TP53, respectively. The 2-year OS rates (Table 2) were not significantly different according to PPM1D/TP53 mutation status. In a multivariate analysis of age, sex, International Prognostic Scoring System (IPSS) risk group, and PPM1D mutation status, only age and IPSS risk group were independently associated with OS.
Only two patients with other MDS or sAML with del(5q) had mutated PPM1D, both with co-mutated TP53 and complex karyotype, so the independent prognostic value of mutated PPM1D could not be assessed. Patients with mutated TP53 (with or without mutated PPM1D) had a significantly shorter 2-year OS than patients with wildtype PPM1D and TP53 (p = 0.004 and p < 0.001 for monoallelic and biallelic TP53 mutations, respectively), (Table 2).
Table 2. OS according to mutational status*
|
Del, deletion; MDS, myelodysplastic syndromes; mut, mutation; OS, overall survival; sAML, secondary AML; WHO, World Health Organization; wt, wildtype. |
|
|
Status |
2-year OS, % |
|---|---|
|
WHO 2016 defined del(5q) MDS |
|
|
PPM1Dmut +/− TP53mut (n = 11) |
100 |
|
PPM1Dwt/TP53mut (n = 16) |
100 |
|
PPM1Dwt/TP53wt (n = 137) |
85 |
|
Other MDS or sAML with del(5q) |
|
|
TP53mut, monoallelic +/− PPM1Dmut (n = 9) |
11 |
|
TP53mut, biallelic +/− PPM1Dmut (n = 6) |
0 |
|
PPM1Dwt/TP53wt (n = 37) |
53 |
Effect of lenalidomide treatment
Hematologic response was achieved in 83.1% of patients with WHO 2016 defined del(5q) MDS treated with lenalidomide (n = 65). Treatment response was independent of PPM1D and TP53 mutation status. After a median follow-up of 3.1 years, 61.5% of patients treated with lenalidomide became refractory or had progressed to AML, which was independent of mutation status.
Changes in mutation status after lenalidomide treatment were investigated for 22 patients with MDS and del(5q); five achieved a complete response and 17 developed resistance to lenalidomide and either progressed (n = 7) or transformed to AML (n = 10).
Only one of the five patients achieving a complete response had mutations before treatment, in PPM1D and ASXL1. The variant allele frequency (VAF) decreased after 76 months of lenalidomide treatment from 27.6% to 4.8% for PPM1D and 12.1% to 1.1% for ASXL1.
For the 17 patients that progressed/transformed to AML, prevalence of mutated PPM1D and/or TP53 increased from 29.4% to 52.9% (p = 0.09; Figure 1), and VAF of PPM1D and TP53 mutations increased from 10.2% to 23.3%, and 5.9% to 23.2%, respectively.
Figure 1. Changes in PPM1D and TP53 mutation status after treatment with lenalidomide for patients who progressed or transformed to AML*
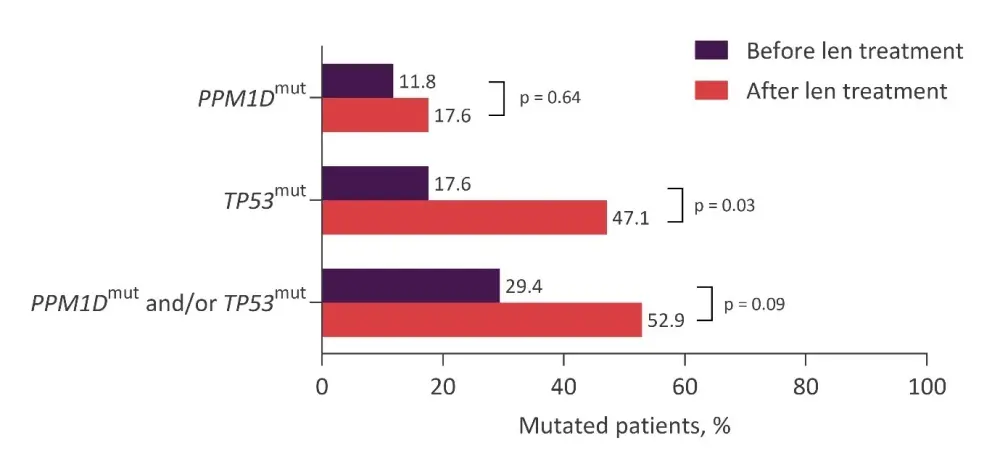
Len, lenalidomide; mut, mutation.
*Data from Panagiota et al.1
Summary
PPM1D and TP53 mutations were present in 5.6% and 15% of patients with MDS or sAML and del(5q), respectively, prior to treatment with lenalidomide. There was no prognostic impact of PPM1D or TP53 mutations in WHO 2016 defined del(5q) MDS, but mutated TP53 was associated with inferior survival in patients with other MDS or sAML with del(5q).
Novel PPM1D and TP53 mutations were acquired in patients who progressed or transformed to AML and further studies are warranted to determine whether assessment of clonal change will help to identify patients at risk of inferior outcomes.
References
Please indicate your level of agreement with the following statements:
The content was clear and easy to understand
The content addressed the learning objectives
The content was relevant to my practice
I will change my clinical practice as a result of this content

