All content on this site is intended for healthcare professionals only. By acknowledging this message and accessing the information on this website you are confirming that you are a Healthcare Professional. If you are a patient or carer, please visit the MDS Alliance.
The mds Hub website uses a third-party service provided by Google that dynamically translates web content. Translations are machine generated, so may not be an exact or complete translation, and the mds Hub cannot guarantee the accuracy of translated content. The mds and its employees will not be liable for any direct, indirect, or consequential damages (even if foreseeable) resulting from use of the Google Translate feature. For further support with Google Translate, visit Google Translate Help.
Now you can support HCPs in making informed decisions for their patients
Your contribution helps us continuously deliver expertly curated content to HCPs worldwide. You will also have the opportunity to make a content suggestion for consideration and receive updates on the impact contributions are making to our content.
Find out more
Create an account and access these new features:
Bookmark content to read later
Select your specific areas of interest
View MDS content recommended for you
Personalized prediction model for risk stratification in patients with MDS
Patients with myelodysplastic syndromes (MDS) present with heterogenous clinical outcomes, with some patients living longer while others dying within a few months of diagnosis. Accurately predicting outcomes in these patients may help in identifying suitable therapies. The International Prognostic Scoring System (IPSS) and the revised IPSS (IPSS-R) are the most used prognostic models in clinical practice and the addition of molecular data to these scorings systems allows upstaging and downstaging of patients into more appropriate risk categories. However, the increment in improving the accuracy of the model is modest and may under- or over-estimate survival in patients due to the models being developed in untreated patients.
Here, we summarize the findings of a cohort study on the development and validation of a prediction model providing a personalized prognosis for patients with MDS published by Nazha and colleagues1 in the Journal of Clinical Oncology.
Methods
A cohort study with a training cohort of 1,471 patients (Cleveland Clinic, n = 528; and Munich Leukemia, n = 943) with MDS, including their comprehensively annotated clinical and molecular data, analyzed using machine learning technique. A random survival algorithm was used to build a prognostic model, and validated using external cohorts (Moffitt Cancer, S1117, and transplant cohort).
- Twenty-four genes most reported in genomic panels were included for DNA sequencing and analysis.
- Harrell concordance index (c-index) was used to establish and compare the accuracy of the proposed prediction model with other existing models.
- A Bonferroni correction was used to identify significant mutations, and a p value of < 0.002 was considered significant.
Results
Baseline characteristics
The median age of patients in the training and validation cohort was 71 years (range, 19−99) and 70 years (range, 20−92), respectively. Cytogenetic risk categories per IPSS-R included (4% vs 3%) with very good, (72% vs 58%) with good, (13% vs 15%) with intermediate, (4% vs 7%) with poor, and (6% vs 17%) with very poor risk in training and validation cohort, respectively. The clinical and mutational characteristics of both cohorts are summarized in Table 1.
Table 1. Baseline characteristics of training and validation cohort*
|
AML, acute myeloid leukemia; ALC, absolute lymphocyte count; AMC, absolute monocyte count; ANC, absolute neutrophil count; BM, bone marrow; BMT, bone marrow transplant; IPSS, International Prognostic Scoring System; IPSS-R, revised IPSS; MDS, myelodysplastic syndromes; MDS-U, MDS unclassifiable; MLD, multilineage dysplasia; RS, ring sideroblast; SLD, single lineage dysplasia; WBC white blood count; WHO, World Health Organization. |
||
|
Characteristic, % (unless otherwise stated) |
Training cohort |
Validation cohort |
|---|---|---|
|
Transformed to AML |
16 |
22 |
|
Received BMT |
9 |
12 |
|
2016 WHO subtype |
||
|
MDS MLD |
24 |
27 |
|
MDS SLD |
5 |
15 |
|
MDS with excess blasts-1 |
21 |
19 |
|
MDS with excess blasts-2 |
18 |
19 |
|
MDS-SLD/MDS-RS |
24 |
17 |
|
MDS-U |
3 |
2 |
|
Clinical characteristic |
||
|
Median WBC count, k/µL (range) |
4 (1−83) |
6 (0−26) |
|
Median hemoglobin, g/dL (range) |
10 (4−16) |
10 (4−17) |
|
Median platelets, k/µL (range) |
120 (4−975) |
113 (7−1,240) |
|
Median ANC count, k/µL (range) |
2 (0−65) |
2 (0−8) |
|
Median AMC count, k/µL (range) |
2 (0−7) |
0 (0−2) |
|
Median ALC count, k/µL (range) |
5 (0−62) |
1 (0−6) |
|
Median BM blast percentage (range) |
4 (0−19) |
3 (0−19) |
|
Median peripheral blood blast percentage (range) |
0 (0−15) |
0 (0−17) |
|
IPSS risk category |
||
|
Low |
27 |
30 |
|
Intermediate-1 |
43 |
37 |
|
Intermediate-2 |
19 |
23 |
|
High |
5 |
8 |
|
IPSS-R risk category |
||
|
Very low |
22 |
17 |
|
Low |
29 |
31 |
|
Intermediate |
23 |
21 |
|
High |
13 |
12 |
|
Very high |
6 |
18 |
Molecular landscape of MDS
- A minimum of one mutation was identified in 78% of patients in the training cohort with a median of two mutations (range, 0−8) per sample, consistent with previous studies.
- Gene mutations for TP53 and complex karyotype, TP53 and chromosome 7 abnormalities, and STAG2 or RUNX1 and ASXL1 showed a strong correlation. SF3B1 mutations were mutually exclusive with TP53 mutations, complex karyotype, chromosome 7 abnormalities, and ASXL1/SRSF2/U2AF1.
Impact of variables on OS
- Impact of mutations that were either significantly negative or positive in the univariate analysis are shown in Figure 1.
- In the multivariate analysis, mutations fluctuated depending on the variables. For example, only mutations in ASXL1, EZH2, KRAS, NRAS, RAD21, SF3B1, and TP53 were significant when only significant mutations in univariate analysis were added, whereas more mutations—ASXl1, CBL, EZH2, NRAS, RA21, SF3B1, TET2, and TP53—were significant when added to all 24 genes.
- EZH2 was the only mutation to demonstrate a negative impact on OS when adjusted for age, IPSS-R, and Bonferroni correction, with variant allele frequency (VAF) as a continuous variable (hazard ratio [HR] 6; 95% CI, 2−19; p < 0.001).
Figure 1. Impact of mutations on OS in univariate analysis*
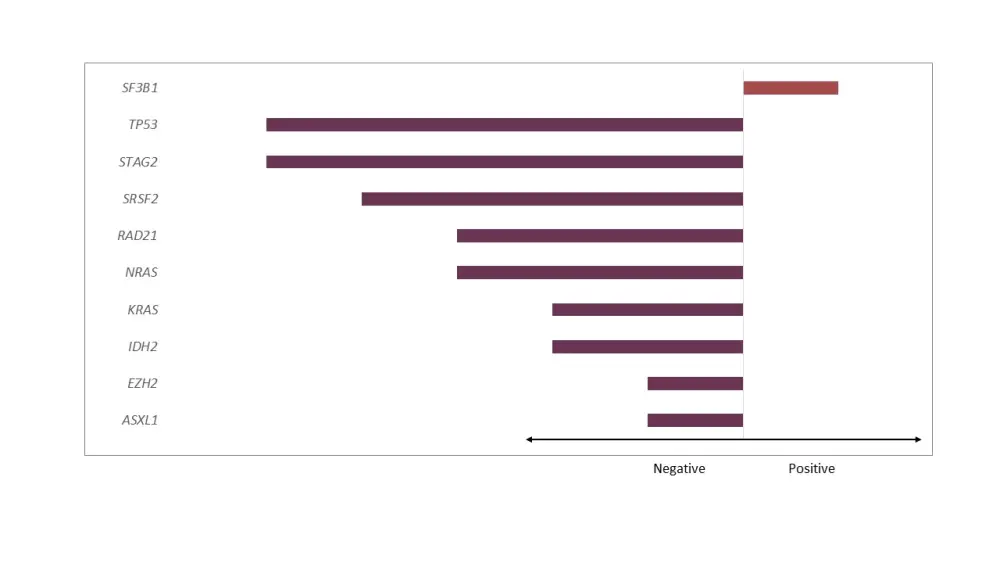
*Adapted from Nazha et al.1
- The most and least important variables having an impact on OS are shown in Figure 2.
- The number of mutations had a significant impact on OS (HR 1; 95% CI, 1−1; p < 0.001) and remained significant irrespective of age and IPSS-R categories.
Figure 2. Impact of variables on OS*
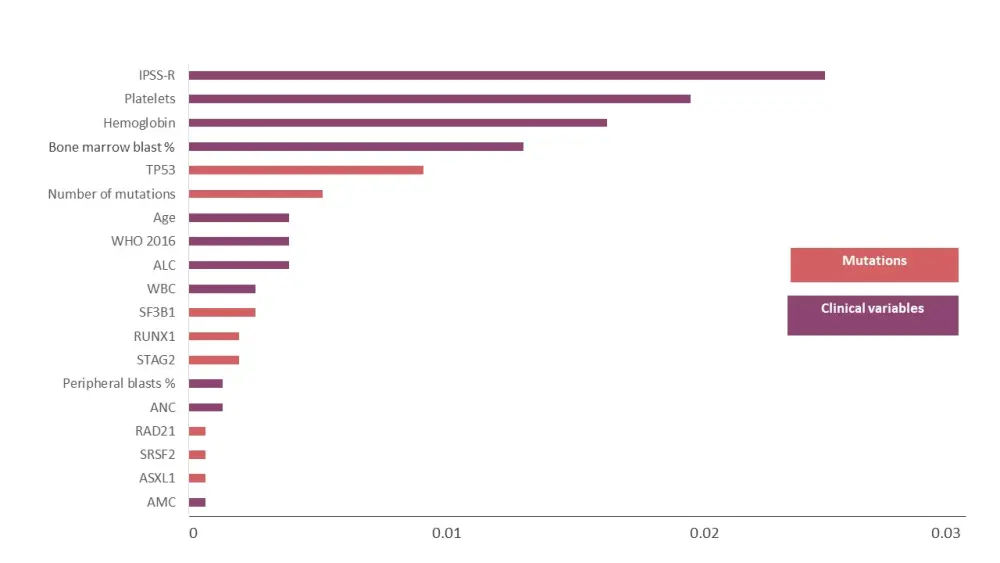
ALC, absolute lymphocyte count; AMC, absolute monocyte count; ANC, absolute neutrophil count; IPSS-R, revised International Prognostic Scoring System; WBC white blood count; WHO, World Health Organization.
*Adapted from Nazha et al.1
Impact of mutations on AML transformation
- Impact on acute myeloid leukemia (AML) transformation also fluctuated depending on the variables included in the multivariate analysis. For example, only mutations in ASXL1, IDH2, PHF6, PTPN11, RAD21, RUNX1, SF3B1, STAG2, and TP53 were significant when only significant mutations in univariate analysis were added, whereas more mutations—ASXl1, IDH2, RAD21, RUNX1, SF3B1, SRSF2, STAG2, and TP53—were significant when all 24 genes were added.
- Mutations in ASXL1 (HR 1; 95% CI, 1−1; p < 0.001), NPM1 (HR 12; 95% CI, 1−103; p = 0.023), RUNX1 (HR 3; 95% CI, 0−36; p = 0.019), and TET2 (HR 5; 95% CI, 1−16; p = 0.013) led to a higher probability of AML transformation with VAF (continuous variable).
- The number of mutations had a significant impact on AML transformation (HR 2; 95% CI, 1−2; p < 0.001) and remained significant irrespective of age and IPSS-R categories.
- The impact was retained even after removal of SF3B1 and TP53 mutations (OS and leukemia-free survival, p < 0.001 in each) for both OS and AML transformation.
Personalized prediction model and its clinical implications
- Cross-validation model showed the c-index score of 0.74 vs 0.71 for OS, 0.81 vs 0.84 for AML transformation and 0.66 vs 0.67 for IPSS-R in the training and validation cohort, respectively.
- Patients with a median OS < 18 months and survival probability of < 50% were considered as higher-risk patients.
- 20% and 16% of the patients in the training and validation cohort, respectively, were identified as higher-risk groups.
- Patients identified as lower-risk per IPSS, and IPSS-R were 48% vs 39%, and 35% vs 45%, in the training and validation cohort, respectively.
Validations of the new model
- The c-index score was 0.68 vs 0.57 for IPSS in the new prediction model and validation S1117 cohort, respectively. In total, 64% (37/58) with higher risk per IPSS were downstaged, and 18% (3/17) were upstaged to a higher risk category using the new prediction model.
- The median OS for the downstaged patients was 26 months vs 16 months in patients without changes in their risk category (p = 0.33).
- The median OS for patients identified as higher risk (new prediction model and IPSS) was 13 months in patients who received azacitidine vs 26 months in patients who received azacitidine plus lenalidomide, and 15 months in patients who received azacitidine plus vorinostat.
- The c-index score was 0.74 vs 0.66 vs 0.64 for IPSS and 0.71 vs 0.68 vs 0.68 for IPSS-R in the new prediction model, at sample 1 and sample 2 from the paired cohort, respectively.
- The c-index score was 0.62 vs 0.58 and 0.60 for the IPSS-R and IPSS in the new prediction model and transplant cohort.
Conclusion
This cohort study developing and validating a prognostic model using clinical and mutational data to estimate the risk of death or progression to AML demonstrated that the model was significantly better than the IPSS and IPSS-R scoring systems. The new prediction model demonstrated reproducibility, generalizability, and stability over time including its ability to be used as a stand-alone model or in conjunction with the IPSS/IPSS-R systems to improve their accuracy. The model also showed the capability to upstage and downstage patients into more appropriate risk categories.
References
Please indicate your level of agreement with the following statements:
The content was clear and easy to understand
The content addressed the learning objectives
The content was relevant to my practice
I will change my clinical practice as a result of this content

